Launch on Feb 10th 2026
〰️
Attend webinar to learn more
Launch on Feb 10th 2026 〰️ Attend webinar to learn more
cfDNA methyl-seq and fragmentomics
with SRSLY® MehtylPLUS
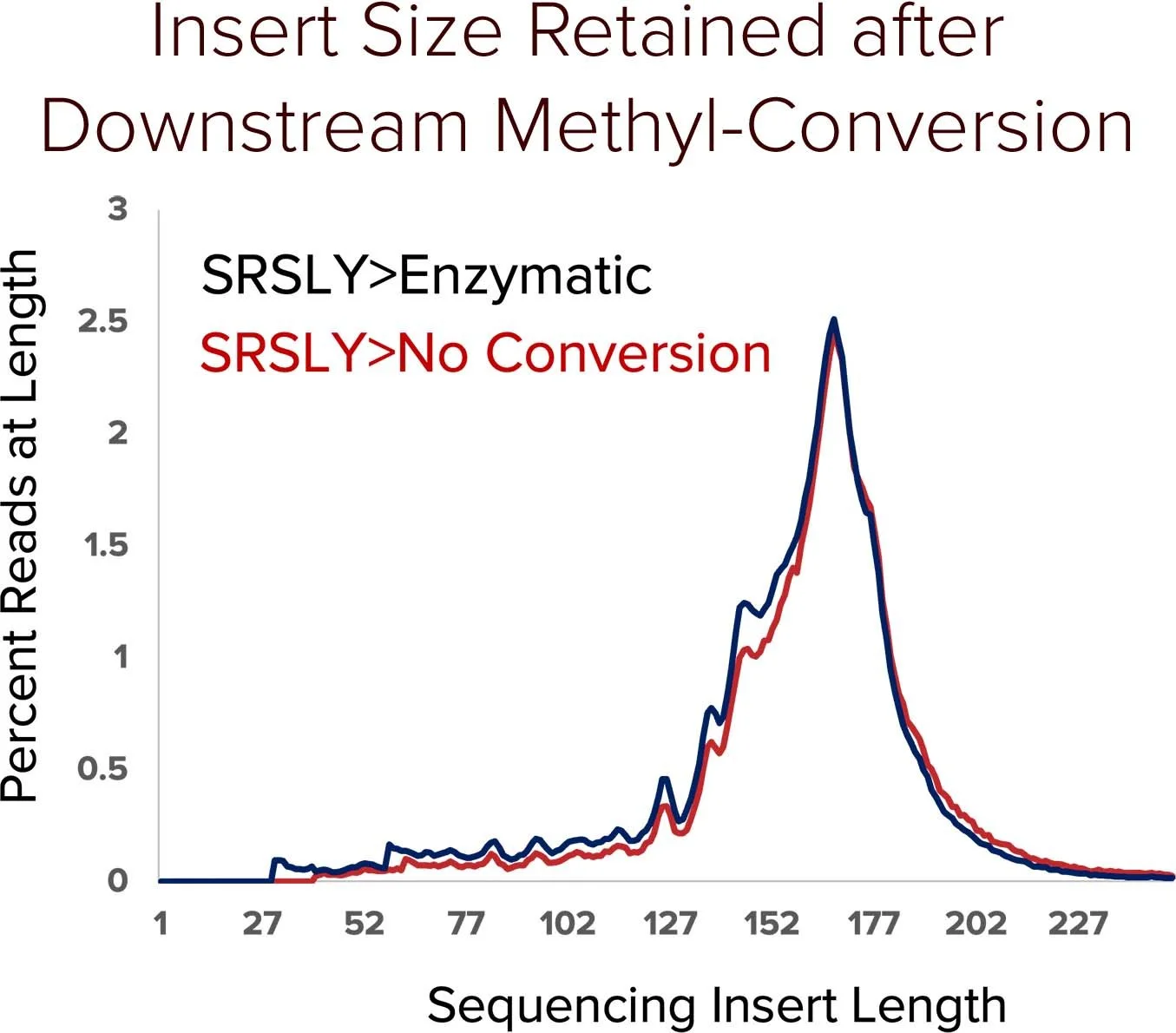
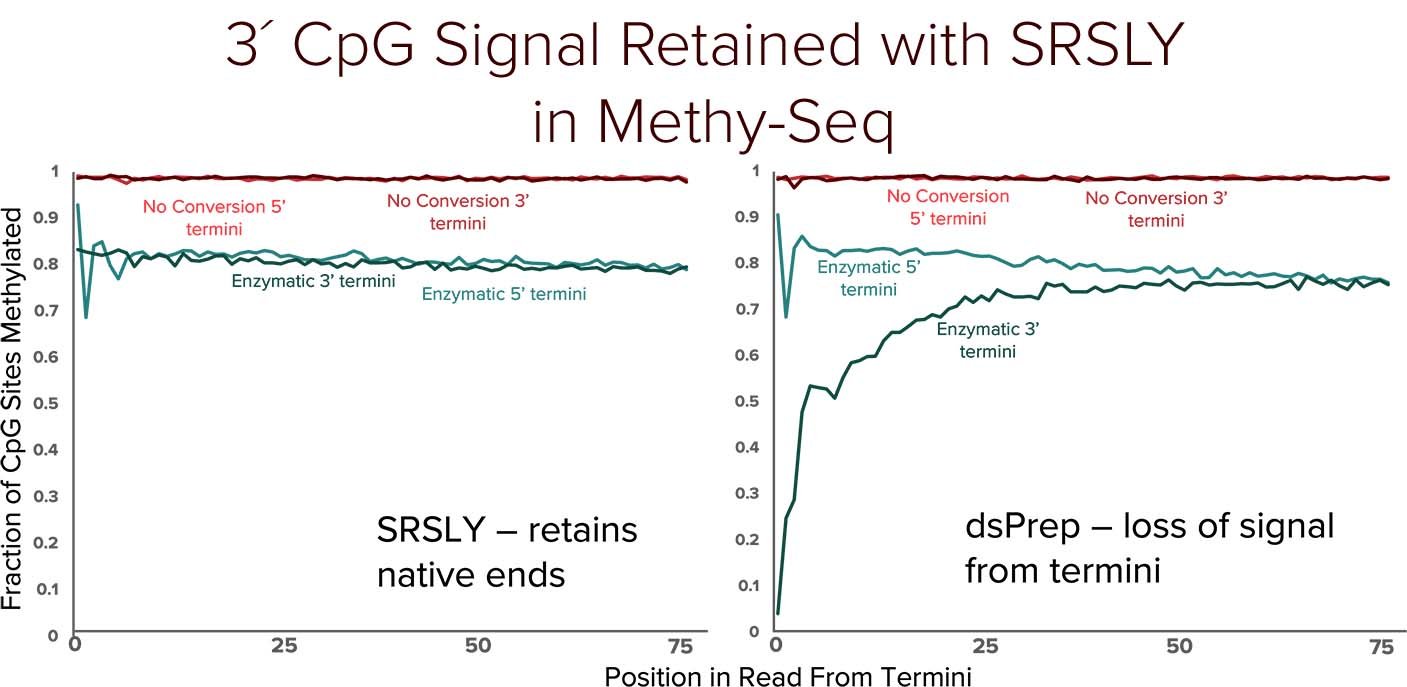
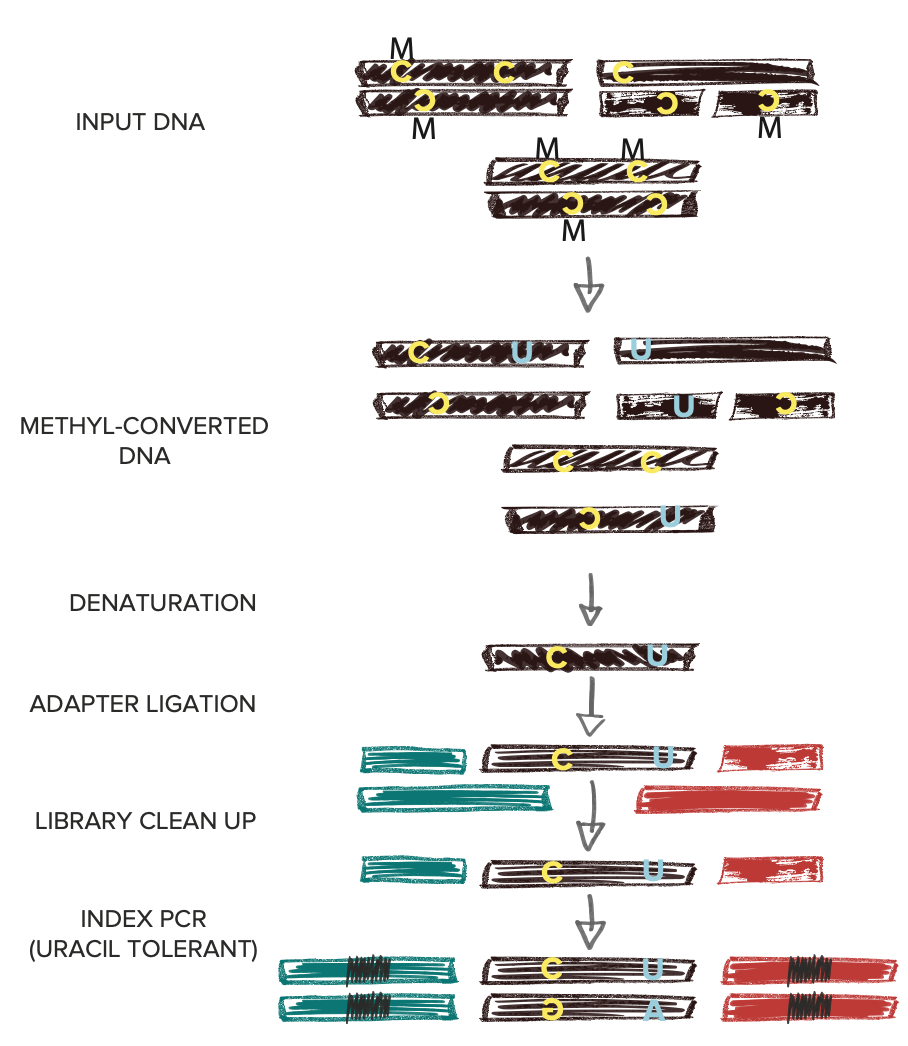
Traditional bisulfite based methyl conversion methods result in spurious fragmentation of cfDNA and double-stranded library preparation methods (dsPreps) lose critical information at cfDNA native termini.
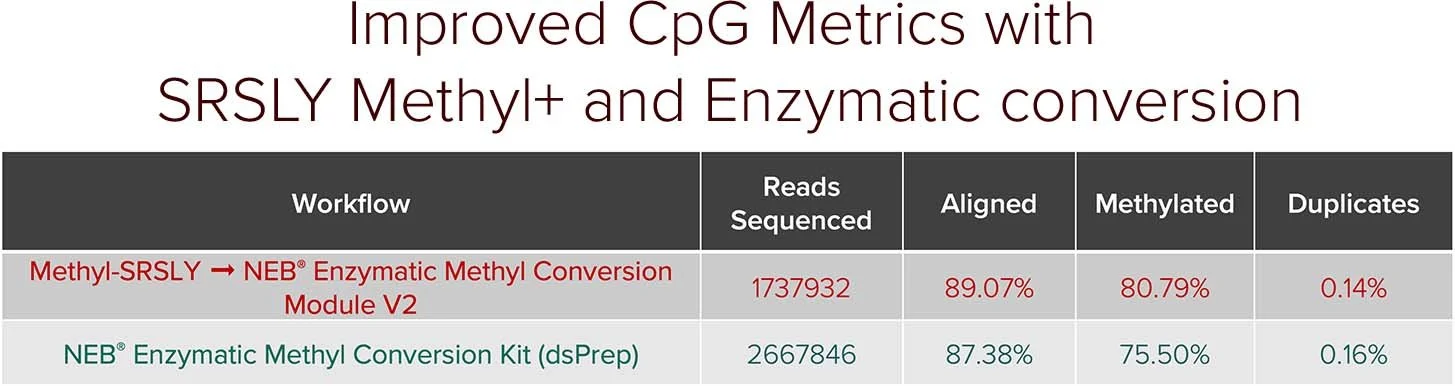
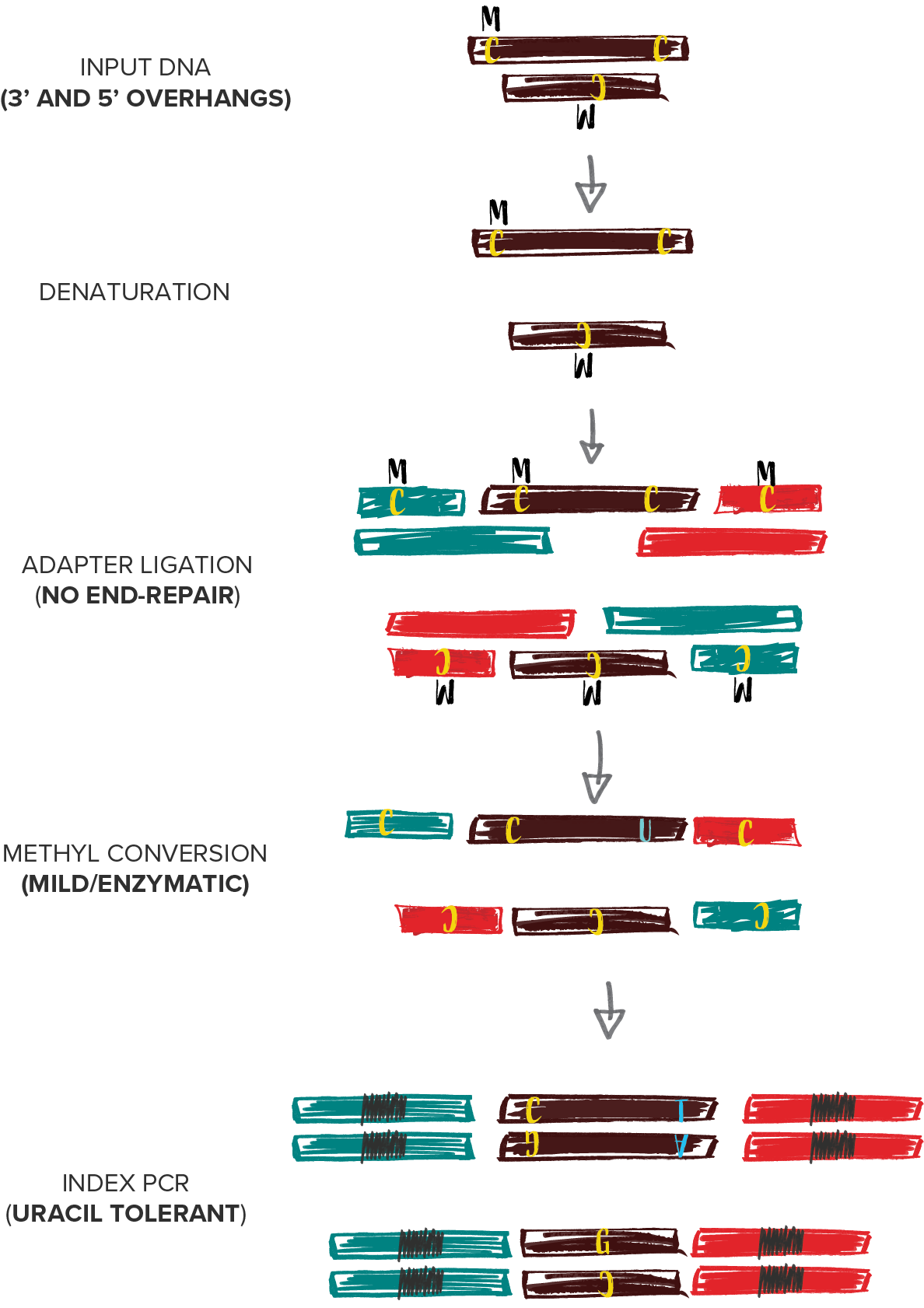
When combined with SRSLY library preparation followed by enzymatic methyl-conversion - captures both fragmentomics and methylation signals along the entire cfDNA fragment. By enabling robust capture of cfDNA methylation patterns and fragmentomic features, SRSLY™ supports early cancer detection research and advanced epigenetic biomarker discovery.
Use our SRSLY MethylPlus kit with Methylated Adapters and Uracil-tolerant Polymerase for Methyl-Seq with cfDNA (250pg - 50ng) and other inputs where native termini retention is critical.
key data highlights
srsly sightings:
Check out the latest publications and preprints that cite SRSLY or our original technology paper for Methyl-Seq applications