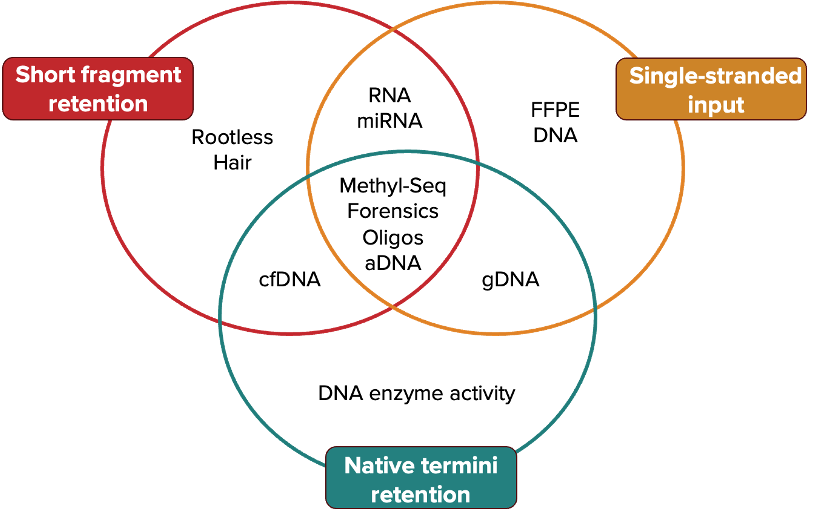
One kit many applications - Click on each To learn more.
cfDNA | FFPE-DNA | MEthyl-seq | cfdna methyl-seq | Ancient DNA | OLIGO QC | RNA-Seq
KEY features
Captures nicked, ssDNA and damaged DNA
Retains native ends
Captures short DNA between 20-50bp
Generates libraries of higher complexity for the same input amount
Higher concordance between expected and observed variant calls, especially for degraded inputs (DIN 3)
Automation friendly, compatible with alternative sequencers
PRODUCT SPECIFICATIONS
Sequence-ready library in under 3 hours
Few PCR cycles required - reduce sequence biases
Compatible with multiple input types
Complete kit with reagents for adapter ligation, indexing PCR and magnetics beads for purification
Optional: ForShear for Enzymatic Shearing for application with FFPE
Optional: UMI-Add on for rare-variant analysis
WHY SRSLY?
As you know, degraded DNA is a very common challenge in NGS workflows. A variety of sample types yield low quality DNA – FFPE tissue, liquid biopsy and rootless hair. These DNA fragments could be short (<100 bp), damaged, nicked or single-stranded and in low amounts (< 1ng). Most library preparation methods are unable to capture all the DNA molecules from such sources.
Launched in 2019, SRSLY library kit from ClaretBio is a single-stranded library preparation method that offers a simple, fast, easily automatable way to convert DNA from challenging inputs into sequencing libraries of high complexity. We have developed additional modules ForShear™, rNONE™ and UMI Add-on to expand the application of our workflow to a broader range of sample types.
Tired of switching NGS kits based on sample quality or quantity? Let us take the guesswork out of it, SRSLY!